funOmics: Aggregating Omics Data into Higher-Level Functional Representations
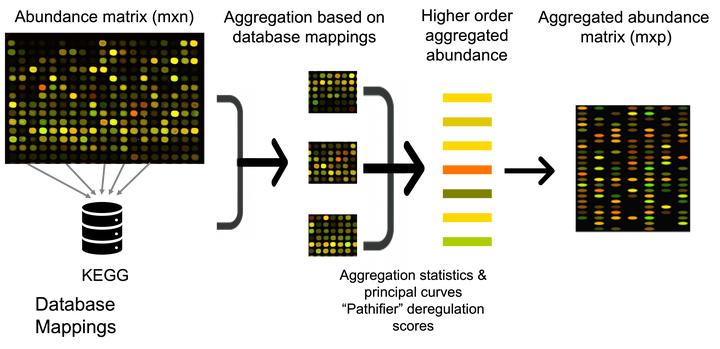 Image credit: From my PhD Thesis
Image credit: From my PhD Thesis
Abstract
The funOmics R package is a collection of functions for aggregating omics data into higher-level functional representations such as pathways, protein complexes, and cellular locations. The package provides a tool for aggregating omics data from high-throughput experiments (e.g. transcriptomics, metabolomics, proteomics) into higher-level functional activity scores that can then be used for further analysis and modeling. The package provides different pooling operators, such as aggregation statistics (mean, median, standard deviation, min, max), dimension-reduction derived scores (pca, nmf, mds, pathifier), or test statistics (t-test, Wilcoxon test, Kolmogorov–Smirnov test) with options for adjusting parameters and settings to suit specific research questions and data types. The package is also well-documented, with detailed descriptions of each function and an example of usage.